Connectome Generation¶
by Lawrence Binding
Estimated Time
20 minutes
Now we have our whole phantom tractogram! Now lets learn how to generate our very own connectome.
Atlas¶
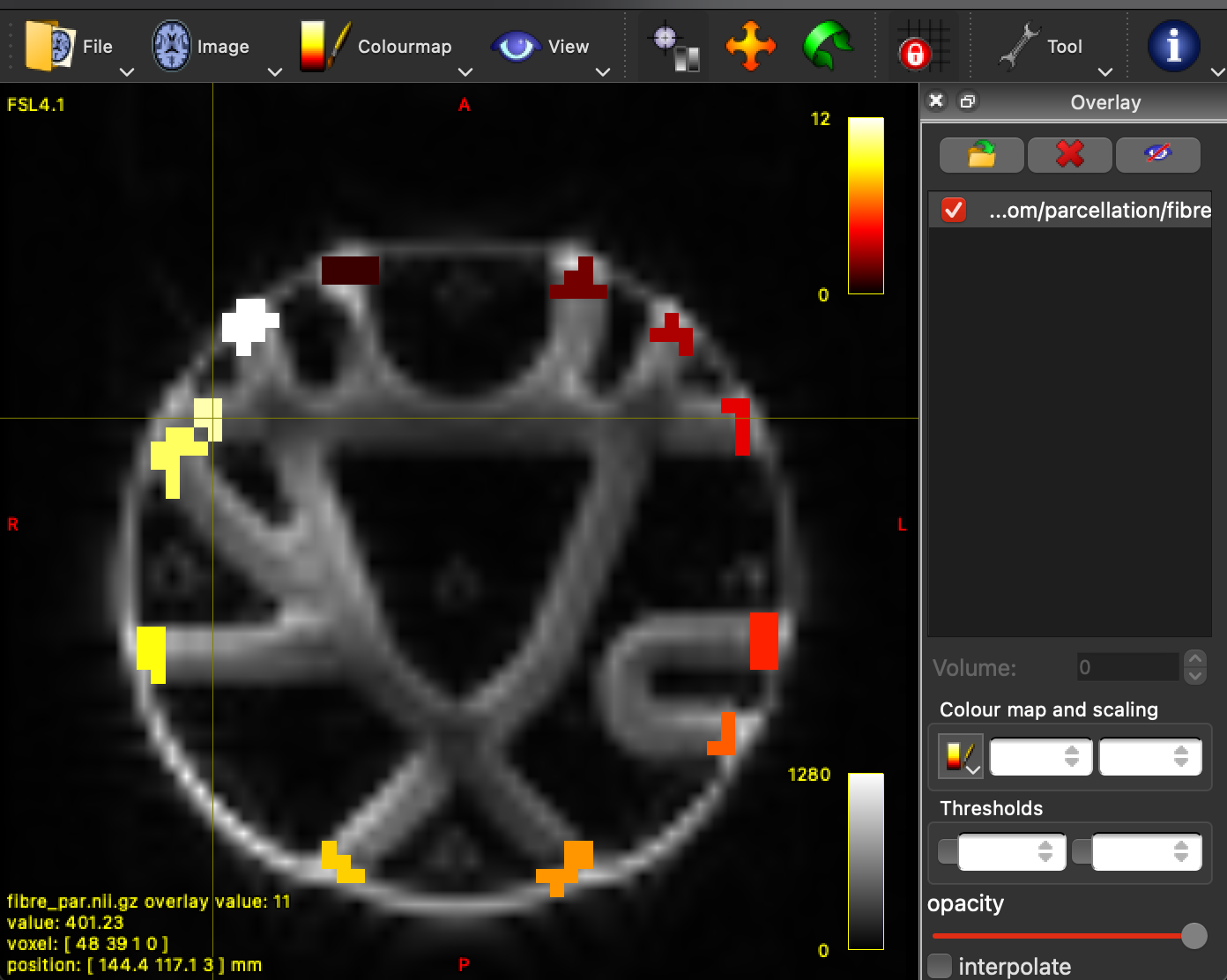
To generate our connectome, we need to have an atlas. A cortical atlas typically contain the cortex split up into functional or anatomical regions. Each region has a unique number assigned to it. There are hundreds of different types of atlas out there, the decision on which one to use will largely depend on what you intend to measure. Thankfully, we don’t have that much decision when it comes to the fibrecup phantom. Located in the drive folder is a file called “fibre_parc.nii.gz”. Open it in overlay, and disable interpolation (located in: tools > overlay, bottom of the panel). Hover over each region and at the bottom left it tells you the number of each region its assigned to.
mrview fibrecup_denoise_gibbs_preproc_biasCorr.mif -overlay.load fibre_parc.nii.gz
Connectome Generation¶
So let's move onto generating our connectome. We can do so using the tck2connectome command:
tck2connectome -symmetric -zero_diagonal tcks_10k.tck fibre_parc.nii.gz MRconnectome.csv
-symmetric is asking it to return a symmetrical matrix
-zero_diagonal is asking it to not return results connecting a parcel to itself.
If you downloaded your tractography result from yesterday's python exercise, you can also use this to generate a connectome to compare with the one from mrtrix. (We have also included this in the google drive folder).
tck2connectome -symmetric -zero_diagonal python_1k_streamlines.tck fibre_parc.nii.gz PYconnectome.csv
Connectome inspection¶
Open up the CSV sheet in Excel. There should be a square array of numbers and lots of zeros. The numbers represent the number of streamlines connecting each pair of regions in the atlas. You can see that the connectome is symmetric about the diagonal.
Further reading
ATLAS INFO: https://neuroimaging-core-docs.readthedocs.io/en/latest/pages/atlases.html
https://mrtrix.readthedocs.io/en/latest/reference/commands/tck2connectome.html